 Ubiquitin is a highly conserved protein 76 amino acids in length. Its attachment, known as ubiquitination, endows different fates onto the modified protein, ranging from degradation via the proteasome to changes in subcellular location or activity. Ubiquitination of a substrate protein and its decoding into a signal, involves a sequential cascade of enzymatic activities, which can be subdivided into ‘writers’, ‘readers’, and ‘erasers’.
Ubiquitin is a highly conserved protein 76 amino acids in length. Its attachment, known as ubiquitination, endows different fates onto the modified protein, ranging from degradation via the proteasome to changes in subcellular location or activity. Ubiquitination of a substrate protein and its decoding into a signal, involves a sequential cascade of enzymatic activities, which can be subdivided into ‘writers’, ‘readers’, and ‘erasers’.
Ubiquitination is in volved in the orchestration of cellular responses. Some examples inlcude the degradation of repressors by the proteasome to activate transcription in response to hormone perception, or the internalization and transport of activated receptors into the vacuole to dampen cellular signalling.
The overarching interest of our research is to elucidate the molecular basis of ubiquitination, as well as the functions of ubiquitin signalling in the regulation of cellular responses and the maintenance of homeostasis.
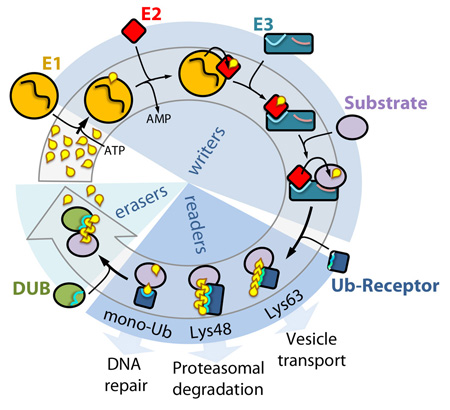
The ubiquitin cycle
Writers: in the initial step, the ubiquitin-activating enzyme (E1) forms a thioester linkage with ubiquitin with consumption of ATP. Next, ubiquitin is passed on to a ubiquitinconjugating enzyme (E2) in a transesterification reaction. The E2–ubiquitin conjugate associates with the ubiquitin ligase (E3), which facilitates the transfer of ubiquitin from the E2 onto an available lysine residue on the substrate. Pairing of the E3 with a specific E2 determines the type of ubiquitination. Substrates can be mono- or poly-monoubiquitinated, or ubiquitins can be attached to one another via one of its seven lysines or Met1. Notably, E3s frequently autoubiquitinate (not shown). Readers: ubiquitination is recognized by ubiquitin receptors, which are able to discriminate between the different types of attachment and mediate specific downstream responses. Erasers: ubiquitin moieties are effectively cleaved of by deubiquitinating (DUB) enzymes, which display catalytic preferences for specific types of ubiquitination.